Faculty Profile
 Dinler Amaral Antunes
Dinler Amaral Antunes
Assistant Professor, Computational Biology
Department of Biology and Biochemistry
Research Division: Biochemistry (Primary)
Office: Science & Engineering Research Center (SERC), 3007
Contact: dinler@uh.edu
Education: D.S., Federal University of Rio Grande do Sul (UFRGS, Brazil); M.S., UFRGS; B.S, UFRGS
Google Scholar Profile
Website
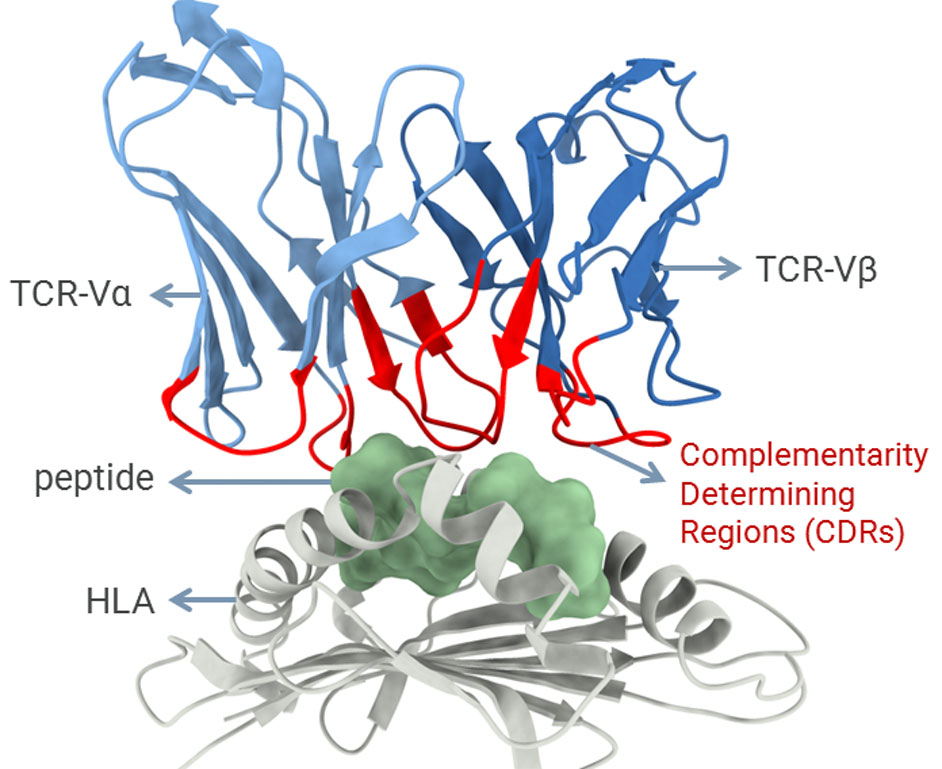
Dr. Dinler Amaral Antunes’ research group uses structural bioinformatics methods, such as molecular modeling, molecular docking and molecular dynamics, to investigate protein-ligand interactions with relevant biomedical applications. Since they are constantly pushing the limits of what can be done with available tools, his group is also actively adapting and developing new computational methods to address specific biological problems.
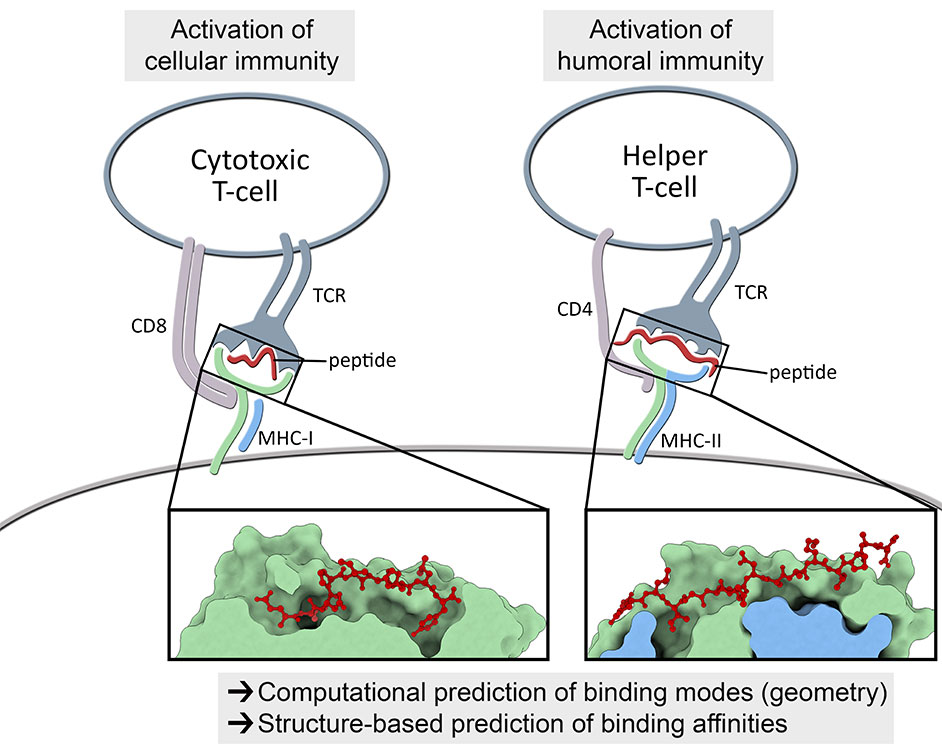
In addition to collaborative projects involving broader biomedical applications (e.g., drug discovery), his lab has a particular focus on studying the mechanisms involved in cellular immunity. This type of adaptive immunity is mediated by T-cell lymphocytes and is key for immunological responses targeting both viruses and cancer cells. T-cells can recognize pieces of proteins (i.e., peptides) displayed at the surface of other cells by a family of receptors known as human leukocyte antigens (HLAs). The recognition of peptide-HLA complexes is mediated by the complementarity determining regions (CDRs) of the T-cell receptor (TCR), representing a central step for the activation of T-cells and the development of cellular immunity.
Understanding the molecular features driving the affinity and specificity of the TCR/pHLA interaction can create new opportunities for biomedical applications across different fields, including antiviral vaccine development, cancer immunotherapy, and the treatment of autoimmune diseases. Over the past decade, Dr. Antunes has developed several tools to enable the computational modeling and structural analysis of peptide-HLA complexes. Now, he wants to combine these computational methods with data from high throughput molecular biology and proteomics approaches, to improve the safety and the efficacy of future T-cell-based immunotherapies.
More at Antunes Lab Research
- J. R. Abella, D. A. Antunes, K. Jackson, G. Lizée, C. Clementi, and L. E. Kavraki. Markov state modeling reveals alternative unbinding pathways for peptide–mhc complexes. Proceedings of the National Academy of Sciences (PNAS), 117(48):30610–30618, Dec 2020 (PDF)
- D. A. Antunes, J. R. Abella, S. Hall-Swan, D. Devaurs, A. Conev, M. Moll, G. Lizée, and L. E. Kavraki. HLA-Arena: a customizable environment for the structural modeling and analysis of peptide-HLA complexes for cancer immunotherapy. JCO Clinical Cancer Informatics, 4:623–636, July 2020 (PDF)
- D. A. Antunes, J. R. Abella, D. Devaurs, M. M. Rigo, and L. E. Kavraki. Structure-based methods for binding mode and binding affinity prediction for peptide-MHC complexes. Curr. Top. Med. Chem., 18(26):2239–2255, 2018 (PDF)
- D. A. Antunes, M. M. Rigo, M. V. Freitas, M. F. A. Mendes, M. Sinigaglia, G. Lizée, L. E. Kavraki, L. K. Selin, M. Cornberg, and G. F. Vieira. Interpreting T-cell cross-reactivity through structure: Implications for TCR-based cancer immunotherapy. Front. Immunol., 8:1210, 2017 (PDF)
Honors & Awards:
- 2020, Immuno-Oncology Young Investigators’ Forum (IOYIF) Ph.D. Postdoc Award (M. D. Anderson & SITC)
- 2020, SCI Gold Oral Presentation Award at the 6th Annual SCI Summer Research Colloquium (Rice University)
- 2019, Selected with Honorable Mention to the Rice University School of Engineering’s Future Faculty Fellows Program
- 2019-2020, Fellowship from the Computational Cancer Biology Training Program (CCBTP)
- 2017, Best Poster Award, 22nd Annual Sealy Center for Structural Biology & Molecular Biophysics Symposium
- 2016, Research Travel Award to attend to the Immune Epitope Database (IEDB) Workshop (San Diego, CA)
- 2016, Best Poster Award, Development of Novel Therapies Through Fragment Based Drug Discovery Meeting
- 2014-2015, Postdoctoral Fellowship through the Brazilian Scientific Mobility Program (Ciências sem Fronteiras/CNPq)
- 2012, Research Travel Award to visit the Hannover Medical School (Hannover, Germany)
- 2012, Best Poster Award,International Society for Computational Biology Latin American Regional Meeting
- 2011-2014, D.Sc. Fellowship, National Council for Scientific and Technological Development (CNPq)
- 2009-2011, M.Sc. Fellowship, National Council for Scientific and Technological Development (CNPq)
- 2006-2008, Scientific Initiation Fellowship, National Council for Scientific and Technological Development (CNPq)
Organizations, Outreach, Boards, Memberships
- Associate Editor, Frontiers in Bioinformatics, Protein Bioinformatics Section, 2021-present
- Member, American Association for the Advancement of Science (AAAS), 2020-present
- Member, International Society for Computational Biology (ISCB), 2019-present
- Coordinator of the Houston Chapter of an association of Brazilian researchers (PUB Houston), 2016-2020
- Regular Contributor to the Blog of the Brazilian Society for Immunology (SBI), 2014-present