BIOLOGICAL FUNCTIONS OF NUCLEOTIDES
·
Precursors
of DNA and RNA.
·
Activated
intermediates in many biosyntheses: e.g UDP-glucose ® glycogen, CDP-diacylglycerol ® phosphoglycerides, S-adenosylmathionine as methyl donor, etc.
·
Nucleotside triphosphates, especially ATP, as
the universal currency of energy in biological systems.
·
Adenine
nucleotides are components of the coenzymes, NAD(P)+, FAD, and CoA.
·
Metabolic
regulators: (a)
c-AMP is the mediator of hormonal actions; (b)
ATP-dependent protein phosphorylation - activates phosphorylase and inactivates
glycogen synthase; (c) adenylation of a Tyr of bacterial glutamine synthetase -
more sensitive to feedback inhibition and less active; (d) allosteric regulator - glycogen phosphorylase activated by ATP
and inactivated by AMP.
NOMENCLATURE AND
MOLECULAR STRUCTURES
See Table 3-1
SYNTHESIS OF
PURINE RIBONUCLEOTIDES
General
Synthetic Strategy
1. Fisrt, build up attachment on
the a-D-ribose.
2. Next, cyclize the attachment to
form the purine ring.
Important
Features
·
Important
initial findings from the identification of the labeling pattern of uric acid
isolated from pigeons fed with various isotopically labeled compounds.
·
Divergent
organisms (such as E. coli, yeast,
pigeon, human) have virtually identical pathways for the biosynthesis of purine
nucleotides.
·
The
initially synthesized purine derivative is inosine monophosphate (IMP).
·
See
figure on p. 788 for the biosynthetic origins of purine ring atoms.
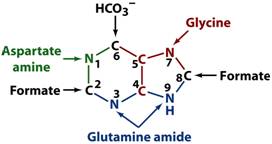
The Pathway for
the Biosynthesis of IMP
·
See
Fig. 22-1.
Synthesis of AMP
and GMP from IMP
·
See
Fig. 22-3.
·
The
synthesis of GMP from IMP requires ATP, whereas the synthesis of AMP from IMP
requires GTP. This is a way to prevent
any excessive synthesis of either AMP or GMP.
·
GMP
is a feedback inhibitor against IMP dehydrogenase.
·
AMP
is a feedback inhibitor against adenylosuccinate
synthetase.
Interconversion of Nucleoside Mono-, Di- and Triphosphate
1. Nucleoside Monophosphate
Kinase:
NMP + ATP D NDP + ADP
2. Nucleoside Diphosphate Kinase:
NDP + ATP D NTP + ADP
Do not discriminate between
ribose and deoxyribose.
3. Adenylate Kinase:
AMP + ATP D 2 ADP
REGULATION OF
PURINE BIOSYNTHESIS
·
See
Fig. 22-4.
·
Ribose
phosphate pyrophosphokinase sensitive to inhibition
by GDP and ADP.
·
Amidophosphoribosyl transferase sensitive to
activation by 5-phosphoribosyl pyrophosphate (PRPP), and inhibition by XMP,
GMP, GDP, GTP, AMP, ADP, and ATP.
·
IMP
dehydrogenase sensitive to inhibition by GMP.
·
Adenylosuccinate synthetase sensitive to
inhibition by AMP.
·
The
synthesis of GMP requires ATP.
·
The
synthesis of AMP requires GTP.
PURINE
DEGRADATION
·
In
animals, all purine nucleotide and deoxynucleotides → Uric acid.
·
Involves
oxidation: Hypoxanthine → Xanthine → Uric acid by Xanthine
Oxidase. (Fig.
22-17)
Fate of Uric
Acid
·
In
humans and other primates: uric acid
excreted in urine.
·
Birds,
terrestrial reptiles, and many insects:
Also excrete uric acid, often at high levels. These organisms do not excrete urea. Moreover, they convert excess amino acid
nitrogen to uric acid via purine biosynthesis.
Gout
·
A
disease: elevated levels of uric acid in
body fluid → deposition of crystals of sodium urate
→
painful arthritic inflammation.
·
Could
result from a number of metabolic insufficiencies, most of which are not well
characterized.
·
One
well understood case: hypoxanthine-guanine
phosphoribosyltransferase deficiency →
PRPP accumulation ® increased synthesis of purine nucleotides → increased uric acid formation.
Hypoxanthine + phosphoribosyl-pyrophosphate → IMP + PPi
(Guanine) (GMP)
·

Allopurinol (a “Competitive” reversible inhibitor for xanthine oxidase) for treatment.
